5080 字
25 分钟
nnUNetv1在自己数据集上训练
本文只讲述nnunetv1的在2D图像的上复现步骤,对于实现细节可以阅读原文和代码!
复现步骤:
1.下载数据集并安装依赖环境:
git clone https://github.com/MIC-DKFZ/nnUNet.git # 下载代码cd nnUNet # 切换目录conda create -n myenv python=3.9 # 注意nnUNetv2需要python>=3.9conda activate myenvpip install nnunetpip install -e . #最后这个点也不能忽略2. 在nnUNet目录下创建文件夹nnUNetFrame,文件夹结构如下:

3. 创建文件
切换到nnUNetFrame文件夹中创建DATASET文件夹,并在DATASET文件夹下创建nnUNet_preprocessed,nnUNet_raw, nnUNet_trained_models文件夹,在文件夹nnUNet_raw,创建nnUNet_cropped_data文件夹和nnUNet_raw_data文件夹,文件夹结构如下:

4. 以linux系统为例,找到.bashrc文件,在末尾添加nnUNet_preprocessed,nnUNet_raw, nnUNet_trained_models的路径,格式如下:
注意:'../'需要替换为本地路径!!!export nnUNet_raw_data_base="../nnUNet/nnUNetFrame/DATASET/nnUNet_raw"export nnUNet_preprocessed="../nnUNet/nnUNetFrame/DATASET/nnUNet_preprocessed"export RESULTS_FOLDER="../nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models"然后关闭.bashrc文件,并在.bashrc文件所在文件目录下运行:
source .bashrc5. 将数据转为nii.gz格式,并生成对应的dataset.json文件:
(1)将原始数据按照如下格式设置:

training为训练集,,testing为测试集。input放置图片,output放置标签。
(2)在nnUNet_raw_data文件夹下创建新的文件夹命名为:Task01_XXX. 01可以修改为任意数字,XXX是任务名,根据自己的任务命名即可;
6. 执行数据转换:
nnUNet_convert_decathlon_task -i Task01_XXX的绝对路径执行完之后会在Task01_XXX同级目录下生成一个文件夹命名为Task001_XXX,示例如下图所示:
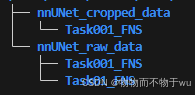
注:如果不清楚原始的文件格式,可以在转换完之后附转为png检查一下是否正确。防止出现了不正确文件导致后续运行报错!
7. 数据预处理
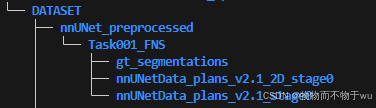
nnUNet_plan_and_preprocess -t 1 --verify_dataset_integrity“1 表示任务代号,即Task001”AI写代码python运行12运行该命令之后会在nnUNet_cropped_data文件中生成命名为Task001_XXX的文件,目录结构如图:
8.训练命令,按顺序运行
CUDA_VISIBLE_DEVICES=1 nnUNet_train 2d nnUNetTrainerV2 Task001_XXX 0 --npzCUDA_VISIBLE_DEVICES=1 nnUNet_train 2d nnUNetTrainerV2 Task001_XXX 1 --npzCUDA_VISIBLE_DEVICES=1 nnUNet_train 2d nnUNetTrainerV2 Task001_XXX 2 --npzCUDA_VISIBLE_DEVICES=1 nnUNet_train 2d nnUNetTrainerV2 Task001_XXX 3 --npzCUDA_VISIBLE_DEVICES=1 nnUNet_train 2d nnUNetTrainerV2 Task001_XXX 4 --npz‘CUDA_VISIBLE_DEVICES=1’ 表示指定GPU训练
‘2d’ 是选用2D Unet模型
‘Task001_XXX’ 表示任务编码,Task001_XXX
‘0,1,2,3,4’ 代表五折交叉验证
9.测试模型:
运行完成五折交叉验证之后可以确定最佳的模型,使用下面的命令进行测试:
nnUNet_find_best_configuration -m 2d -t 001 –strict# 001是任务编号然后会在 nnUNet_trained_models/nnUNet/ensembles/Task001_XXX下生成如下如所示文件:

nnUNet_predict -i FOLDER_WITH_TEST_CASES -o OUTPUT_FOLDER_MODEL1 -tr nnUNetTrainerV2 -ctr nnUNetTrainerV2CascadeFullRes -m 2d -p nnUNetPlansv2.1 -t Task001_XXX# FOLDER_WITH_TEST_CASES 输入文件路径# OUTPUT_FOLDER_MODEL1 输出文件路径# Task001_XXX 预测任务名将上述参数修改为自己的任务,然后运行即可
10. 在输出的路径下会保存预测结果文件
格式为nii.gz格式,需要将其转为png格式,代码如下:
import osimport nibabel as nibimport numpy as npfrom PIL import Image
def convert_nii_to_png(input_folder, output_folder): # 确保输出文件夹存在 os.makedirs(output_folder, exist_ok=True)
# 遍历输入文件夹中的所有文件 for filename in os.listdir(input_folder): if filename.endswith('.nii.gz'): # 构建完整的文件路径 file_path = os.path.join(input_folder, filename) # 读取 NIfTI 文件 nii_image = nib.load(file_path) image_data = nii_image.get_fdata()
# 选择中间的切片 slice_idx = image_data.shape[2] // 2 slice_data = image_data[:, :, slice_idx]
# 转换为8位图像格式 slice_normalized = (slice_data - np.min(slice_data)) / (np.max(slice_data) - np.min(slice_data)) image_8bit = (slice_normalized * 255).astype(np.uint8) image = Image.fromarray(image_8bit)
# 保存图像 output_filename = filename.replace('.nii.gz', '.png') image.save(os.path.join(output_folder, output_filename)) print(f"Converted {filename} to {output_filename}")input_folder = '' # nii.gz文件路径output_folder = '' # png文件路径convert_nii_to_png(input_folder, output_folder)11. 完成!
BraTS数据集转换成MSD数据集
BraTS2019:
import osimport shutilimport jsonimport nibabel as nibimport numpy as npfrom collections import OrderedDict
def convert_brats2019_to_nnunet(brats_root, nnunet_raw_data_base): """ 将BraTS2019数据集转换为nnUNet格式,处理标签映射
Args: brats_root: BraTS2019原始数据根目录路径 nnunet_raw_data_base: nnUNet原始数据基础目录路径 """
# 设置路径 task_name = "Task001_BraTS2019" task_folder = os.path.join(nnunet_raw_data_base, "nnUNet_raw_data", task_name)
# 创建必要的目录 imagesTr_folder = os.path.join(task_folder, "imagesTr") imagesTs_folder = os.path.join(task_folder, "imagesTs") labelsTr_folder = os.path.join(task_folder, "labelsTr") labelsTs_folder = os.path.join(task_folder, "labelsTs")
for folder in [imagesTr_folder, imagesTs_folder, labelsTr_folder, labelsTs_folder]: os.makedirs(folder, exist_ok=True)
# 获取训练数据和测试数据路径 hgg_folder = os.path.join(brats_root, "HGG") lgg_folder = os.path.join(brats_root, "LGG")
# 收集所有训练病例 train_cases = []
# 处理HGG数据 if os.path.exists(hgg_folder): hgg_cases = [d for d in os.listdir(hgg_folder) if os.path.isdir(os.path.join(hgg_folder, d))] for case in hgg_cases: train_cases.append(("HGG", case))
# 处理LGG数据 if os.path.exists(lgg_folder): lgg_cases = [d for d in os.listdir(lgg_folder) if os.path.isdir(os.path.join(lgg_folder, d))] for case in lgg_cases: train_cases.append(("LGG", case))
print(f"找到 {len(train_cases)} 个训练病例")
# 模态映射 modality_mapping = { 'flair': '0000', 't1ce': '0001', 't1': '0002', 't2': '0003' }
def process_label(label_path, output_path): """ 处理BraTS标签,将原始标签映射为连续的标签 BraTS原始标签: 0(背景), 1(坏死/非增强肿瘤), 2(水肿), 4(增强肿瘤) 映射后标签: 0(背景), 1(坏死/非增强肿瘤), 2(水肿), 3(增强肿瘤) """ # 加载标签数据 label_nii = nib.load(label_path) label_data = label_nii.get_fdata().astype(np.uint8)
# 检查原始标签值 unique_labels = np.unique(label_data) print(f"处理 {os.path.basename(label_path)},原始标签值: {unique_labels}")
# 创建新的标签数组 new_label_data = np.zeros_like(label_data)
# 标签映射: 0->0, 1->1, 2->2, 4->3 new_label_data[label_data == 0] = 0 # 背景 new_label_data[label_data == 1] = 1 # 坏死和非增强肿瘤 new_label_data[label_data == 2] = 2 # 水肿 new_label_data[label_data == 4] = 3 # 增强肿瘤
# 检查映射后的标签值 new_unique_labels = np.unique(new_label_data) print(f"映射后标签值: {new_unique_labels}")
# 保存新的标签文件 new_label_nii = nib.Nifti1Image(new_label_data, label_nii.affine, label_nii.header) nib.save(new_label_nii, output_path)
training_cases = [] test_cases = []
for i, (grade, case_name) in enumerate(train_cases): case_folder = os.path.join(brats_root, grade, case_name)
if not os.path.exists(case_folder): print(f"警告: 病例文件夹不存在 {case_folder}") continue
# 检查所有必需的文件是否存在 required_files = { 'flair': f"{case_name}_flair.nii", 't1ce': f"{case_name}_t1ce.nii", 't1': f"{case_name}_t1.nii", 't2': f"{case_name}_t2.nii", 'seg': f"{case_name}_seg.nii" }
# 检查.nii.gz格式 for key, filename in required_files.items(): if not os.path.exists(os.path.join(case_folder, filename)): # 尝试.nii.gz格式 gz_filename = filename + ".gz" if os.path.exists(os.path.join(case_folder, gz_filename)): required_files[key] = gz_filename else: print(f"警告: 文件 {filename} 或 {gz_filename} 不存在于 {case_folder}") break else: # 所有文件都存在,处理这个病例
# 决定这个病例是用于训练还是测试(这里简单地将前80%用于训练) if i < len(train_cases) * 0.8: # 训练数据 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTr_folder, f"{case_name}_{suffix}.nii.gz")
# 如果源文件不是.gz格式,需要压缩 if not src_file.endswith('.gz'): img = nib.load(src_file) nib.save(img, dst_file) else: shutil.copy2(src_file, dst_file)
# 处理分割标签(重要:进行标签映射) src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTr_folder, f"{case_name}.nii.gz") process_label(src_seg, dst_seg)
training_cases.append(case_name)
else: # 测试数据 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTs_folder, f"{case_name}_{suffix}.nii.gz")
if not src_file.endswith('.gz'): img = nib.load(src_file) nib.save(img, dst_file) else: shutil.copy2(src_file, dst_file)
# 测试数据的标签(同样进行标签映射) src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTs_folder, f"{case_name}.nii.gz") process_label(src_seg, dst_seg)
test_cases.append(case_name)
print(f"处理完成: {len(training_cases)} 个训练病例, {len(test_cases)} 个测试病例")
# 创建dataset.json文件 dataset_json = OrderedDict() dataset_json['name'] = "BraTS2019" dataset_json['description'] = "Brain Tumor Segmentation Challenge 2019" dataset_json['tensorImageSize'] = "4D" dataset_json['reference'] = "https://www.med.upenn.edu/cbica/brats2019/" dataset_json['licence'] = "see BraTS2019 website" dataset_json['release'] = "1.0"
# 模态信息 dataset_json['modality'] = { "0": "FLAIR", "1": "T1ce", "2": "T1", "3": "T2" }
# 标签信息 - 修正为包含所有必要的标签 dataset_json['labels'] = { "0": "background", "1": "necrotic/non-enhancing tumor", "2": "edema", "3": "enhancing tumor" }
# 训练和测试数据列表 dataset_json['numTraining'] = len(training_cases) dataset_json['numTest'] = len(test_cases)
dataset_json['training'] = [] for case in training_cases: case_dict = { "image": f"./imagesTr/{case}.nii.gz", "label": f"./labelsTr/{case}.nii.gz" } dataset_json['training'].append(case_dict)
dataset_json['test'] = [] for case in test_cases: dataset_json['test'].append(f"./imagesTs/{case}.nii.gz")
# 保存dataset.json json_file_path = os.path.join(task_folder, "dataset.json") with open(json_file_path, 'w') as f: json.dump(dataset_json, f, indent=4)
print(f"dataset.json 已保存到: {json_file_path}") print("数据转换完成!标签已正确映射:0(背景) -> 0, 1(坏死) -> 1, 2(水肿) -> 2, 4(增强肿瘤) -> 3")
return task_folder
# 使用示例if __name__ == "__main__": # 设置路径 brats_root = "/root/autodl-tmp/nnUNet_raw_data_base/BraTS2019" # 您的BraTS2019数据根目录 nnunet_raw_data_base = "/root/autodl-tmp/nnUNet_raw_data_base" # nnUNet原始数据基础目录
# 执行转换 convert_brats2019_to_nnunet(brats_root, nnunet_raw_data_base)BraTS2023
import osimport shutilimport jsonimport nibabel as nibimport numpy as npfrom collections import OrderedDict
def convert_brats2023_to_nnunet(brats_root, nnunet_raw_data_base): """ 将BraTS2023数据集转换为nnUNet格式,保持原始标签不变
Args: brats_root: BraTS2023原始数据根目录路径 nnunet_raw_data_base: nnUNet原始数据基础目录路径 """
# 错误记录列表 errors = []
# 设置路径 task_name = "Task001_BraTS2023" task_folder = os.path.join(nnunet_raw_data_base, "nnUNet_raw_data", task_name)
# 创建必要的目录 imagesTr_folder = os.path.join(task_folder, "imagesTr") imagesTs_folder = os.path.join(task_folder, "imagesTs") labelsTr_folder = os.path.join(task_folder, "labelsTr") labelsTs_folder = os.path.join(task_folder, "labelsTs")
for folder in [imagesTr_folder, imagesTs_folder, labelsTr_folder, labelsTs_folder]: os.makedirs(folder, exist_ok=True)
# 收集所有病例文件夹 all_cases = [] for item in os.listdir(brats_root): case_path = os.path.join(brats_root, item) if os.path.isdir(case_path) and item.startswith('BraTS-GLI-'): all_cases.append(item)
all_cases.sort() # 确保顺序一致 print(f"找到 {len(all_cases)} 个病例")
# 模态映射 - BraTS2023使用t1c而不是t1ce modality_mapping = { 't1n': '0000', # T1 native (非增强T1) 't1c': '0001', # T1 contrast enhanced (增强T1) 't2f': '0002', # T2 FLAIR 't2w': '0003' # T2 weighted }
def safe_copy_image(src_path, dst_path, case_name, modality): """ 安全地复制图像文件,处理可能的错误 """ try: if not src_path.endswith('.gz'): img = nib.load(src_path) nib.save(img, dst_path) else: shutil.copy2(src_path, dst_path) return True except Exception as e: error_msg = f"复制图像失败 - 病例: {case_name}, 模态: {modality}, 文件: {src_path}, 错误: {str(e)}" print(f"错误: {error_msg}") errors.append(error_msg) return False
def safe_copy_label(src_path, dst_path, case_name): """ 安全地复制标签文件,处理可能的错误 """ try: # 加载标签数据以检查标签值 label_nii = nib.load(src_path) label_data = label_nii.get_fdata().astype(np.uint8)
# 检查原始标签值 unique_labels = np.unique(label_data) print(f"处理 {os.path.basename(src_path)},标签值: {unique_labels}")
# 直接保存,不进行任何修改 if not src_path.endswith('.gz'): # 如果源文件不是.gz格式,保存为.gz格式 nib.save(label_nii, dst_path) else: # 如果已经是.gz格式,直接复制 shutil.copy2(src_path, dst_path) return True except Exception as e: error_msg = f"复制标签失败 - 病例: {case_name}, 文件: {src_path}, 错误: {str(e)}" print(f"错误: {error_msg}") errors.append(error_msg) return False
def check_file_validity(file_path): """ 检查文件是否有效(非空且可读取) """ try: if not os.path.exists(file_path): return False, "文件不存在"
if os.path.getsize(file_path) == 0: return False, "文件为空"
# 尝试加载文件头信息 nib.load(file_path) return True, "文件有效" except Exception as e: return False, f"文件无效: {str(e)}"
training_cases = [] test_cases = [] skipped_cases = []
for i, case_name in enumerate(all_cases): case_folder = os.path.join(brats_root, case_name)
if not os.path.exists(case_folder): error_msg = f"病例文件夹不存在: {case_folder}" print(f"警告: {error_msg}") errors.append(error_msg) skipped_cases.append(case_name) continue
# 构建文件名模式 - 根据BraTS2023的命名规范 base_name = case_name # BraTS-GLI-00000-000
required_files = { 't1n': f"{base_name}-t1n.nii", 't1c': f"{base_name}-t1c.nii", 't2f': f"{base_name}-t2f.nii", 't2w': f"{base_name}-t2w.nii", 'seg': f"{base_name}-seg.nii" }
# 检查文件存在性,支持.nii和.nii.gz格式 files_exist = True invalid_files = []
for key, filename in required_files.items(): file_path = os.path.join(case_folder, filename) gz_file_path = file_path + ".gz"
if os.path.exists(file_path): # 检查文件有效性 is_valid, msg = check_file_validity(file_path) if not is_valid: invalid_files.append(f"{filename}: {msg}") files_exist = False elif os.path.exists(gz_file_path): required_files[key] = filename + ".gz" # 检查文件有效性 is_valid, msg = check_file_validity(gz_file_path) if not is_valid: invalid_files.append(f"{filename}.gz: {msg}") files_exist = False else: error_msg = f"文件缺失 - 病例: {case_name}, 文件: {filename} 或 {filename}.gz" print(f"警告: {error_msg}") errors.append(error_msg) files_exist = False
if invalid_files: for invalid_file in invalid_files: error_msg = f"文件无效 - 病例: {case_name}, {invalid_file}" print(f"警告: {error_msg}") errors.append(error_msg)
if not files_exist: skipped_cases.append(case_name) continue
# 决定这个病例是用于训练还是测试(前80%用于训练) case_success = True
if i < len(all_cases) * 0.8: # 训练数据 print(f"处理训练病例: {case_name}")
# 复制图像文件 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTr_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality): case_success = False
# 复制分割标签 src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTr_folder, f"{case_name}.nii.gz") if not safe_copy_label(src_seg, dst_seg, case_name): case_success = False
if case_success: training_cases.append(case_name) print(f"训练病例 {case_name} 处理成功") else: skipped_cases.append(case_name) print(f"训练病例 {case_name} 处理失败,已跳过")
else: # 测试数据 print(f"处理测试病例: {case_name}")
# 复制图像文件 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTs_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality): case_success = False
# 复制测试数据的标签 src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTs_folder, f"{case_name}.nii.gz") if not safe_copy_label(src_seg, dst_seg, case_name): case_success = False
if case_success: test_cases.append(case_name) print(f"测试病例 {case_name} 处理成功") else: skipped_cases.append(case_name) print(f"测试病例 {case_name} 处理失败,已跳过")
print(f"处理完成: {len(training_cases)} 个训练病例, {len(test_cases)} 个测试病例") print(f"跳过的病例数量: {len(skipped_cases)}")
# 写入错误日志 error_file_path = os.path.join(os.path.dirname(__file__), "error.txt") with open(error_file_path, 'w', encoding='utf-8') as f: f.write(f"BraTS2023数据转换错误报告\n") f.write(f"生成时间: {str(os.path.getctime(error_file_path)) if os.path.exists(error_file_path) else 'N/A'}\n") f.write(f"="*80 + "\n\n") f.write(f"总计处理: {len(all_cases)} 个病例\n") f.write(f"成功处理: {len(training_cases) + len(test_cases)} 个病例\n") f.write(f"跳过病例: {len(skipped_cases)} 个病例\n") f.write(f"错误数量: {len(errors)} 个错误\n\n")
if skipped_cases: f.write("跳过的病例列表:\n") for case in skipped_cases: f.write(f" - {case}\n") f.write("\n")
if errors: f.write("详细错误信息:\n") for i, error in enumerate(errors, 1): f.write(f"{i}. {error}\n") else: f.write("没有发现错误。\n")
print(f"错误日志已保存到: {error_file_path}")
# 只有成功处理的病例数量大于0时才创建dataset.json if len(training_cases) + len(test_cases) > 0: # 创建dataset.json文件 dataset_json = OrderedDict() dataset_json['name'] = "BraTS2023" dataset_json['description'] = "Brain Tumor Segmentation Challenge 2023" dataset_json['tensorImageSize'] = "4D" dataset_json['reference'] = "https://www.synapse.org/#!Synapse:syn51156910" dataset_json['licence'] = "see BraTS2023 website" dataset_json['release'] = "1.0"
# 模态信息 - 更新为BraTS2023的模态 dataset_json['modality'] = { "0": "T1n", # T1 native "1": "T1c", # T1 contrast enhanced "2": "T2f", # T2 FLAIR "3": "T2w" # T2 weighted }
# 标签信息 - 保持BraTS原始标签值 dataset_json['labels'] = { "0": "background", "1": "necrotic/non-enhancing tumor", "2": "edema", "3": "enhancing tumor" # 保持原始标签值3 }
# 训练和测试数据列表 dataset_json['numTraining'] = len(training_cases) dataset_json['numTest'] = len(test_cases)
dataset_json['training'] = [] for case in training_cases: case_dict = { "image": f"./imagesTr/{case}.nii.gz", "label": f"./labelsTr/{case}.nii.gz" } dataset_json['training'].append(case_dict)
dataset_json['test'] = [] for case in test_cases: dataset_json['test'].append(f"./imagesTs/{case}.nii.gz")
# 保存dataset.json json_file_path = os.path.join(task_folder, "dataset.json") with open(json_file_path, 'w') as f: json.dump(dataset_json, f, indent=4)
print(f"dataset.json 已保存到: {json_file_path}") print("BraTS2023数据转换完成!标签保持原始值不变:0(背景), 1(坏死), 2(水肿), 4(增强肿瘤)")
return task_folder else: print("警告: 没有成功处理任何病例,未生成dataset.json文件") return None
# 使用示例if __name__ == "__main__": # 设置路径 brats_root = "/root/autodl-tmp/nnUNet_raw_data_base/BraTS2023" # 您的BraTS2023数据根目录 nnunet_raw_data_base = "/root/autodl-tmp/nnUNet_raw_data_base" # nnUNet原始数据基础目录
# 执行转换 try: result = convert_brats2023_to_nnunet(brats_root, nnunet_raw_data_base) if result: print(f"\n转换成功完成!输出目录: {result}") print("可以继续进行nnUNet的预处理和训练步骤。") else: print("\n转换失败,请查看错误日志了解详细信息。") except Exception as e: print(f"程序执行失败: {str(e)}") # 即使主程序失败,也要记录错误 error_file_path = os.path.join(os.path.dirname(__file__), "error.txt") with open(error_file_path, 'w', encoding='utf-8') as f: f.write(f"程序执行失败: {str(e)}\n") print(f"错误已记录到: {error_file_path}")BraTS2024
import osimport shutilimport jsonimport nibabel as nibimport numpy as npfrom collections import OrderedDict
def convert_brats2024_to_nnunet(brats_root, nnunet_raw_data_base): """ 将BraTS2024数据集转换为nnUNet格式,保持原始标签不变 包含新的切除腔(RC)标签
Args: brats_root: BraTS2024原始数据根目录路径 nnunet_raw_data_base: nnUNet原始数据基础目录路径 """
# 错误记录列表 errors = []
# 设置路径 task_name = "Task001_BraTS2024" task_folder = os.path.join(nnunet_raw_data_base, "nnUNet_raw", task_name)
# 创建必要的目录 imagesTr_folder = os.path.join(task_folder, "imagesTr") imagesTs_folder = os.path.join(task_folder, "imagesTs") labelsTr_folder = os.path.join(task_folder, "labelsTr") labelsTs_folder = os.path.join(task_folder, "labelsTs")
for folder in [imagesTr_folder, imagesTs_folder, labelsTr_folder, labelsTs_folder]: os.makedirs(folder, exist_ok=True)
# 收集所有病例文件夹 all_cases = [] for item in os.listdir(brats_root): case_path = os.path.join(brats_root, item) if os.path.isdir(case_path) and item.startswith('BraTS-'): all_cases.append(item)
all_cases.sort() # 确保顺序一致 print(f"找到 {len(all_cases)} 个病例")
# 模态映射 - BraTS2024模态 modality_mapping = { 't1n': '0000', # T1 native (非增强T1) 't1c': '0001', # T1 contrast enhanced (增强T1) 't2f': '0002', # T2 FLAIR 't2w': '0003' # T2 weighted }
def safe_copy_image(src_path, dst_path, case_name, modality): """ 安全地复制图像文件,处理可能的错误 """ try: if not src_path.endswith('.gz'): img = nib.load(src_path) nib.save(img, dst_path) else: shutil.copy2(src_path, dst_path) return True except Exception as e: error_msg = f"复制图像失败 - 病例: {case_name}, 模态: {modality}, 文件: {src_path}, 错误: {str(e)}" print(f"错误: {error_msg}") errors.append(error_msg) return False
def safe_copy_label(src_path, dst_path, case_name): """ 安全地复制标签文件,处理可能的错误 检查并记录BraTS2024的标签值分布 """ try: # 加载标签数据以检查标签值 label_nii = nib.load(src_path) label_data = label_nii.get_fdata().astype(np.uint8)
# 检查原始标签值 unique_labels = np.unique(label_data) print(f"处理 {os.path.basename(src_path)},标签值: {unique_labels}")
# 验证标签值是否符合BraTS2024规范(0, 1, 2, 3, 4) valid_labels = {0, 1, 2, 3, 4} unexpected_labels = set(unique_labels) - valid_labels if unexpected_labels: warning_msg = f"发现意外标签值 - 病例: {case_name}, 标签值: {unexpected_labels}" print(f"警告: {warning_msg}") errors.append(warning_msg)
# 统计各标签的体素数量 label_counts = {} for label in unique_labels: count = np.sum(label_data == label) label_counts[int(label)] = count
print(f" 标签分布: {label_counts}")
# 直接保存,不进行任何修改 if not src_path.endswith('.gz'): # 如果源文件不是.gz格式,保存为.gz格式 nib.save(label_nii, dst_path) else: # 如果已经是.gz格式,直接复制 shutil.copy2(src_path, dst_path) return True except Exception as e: error_msg = f"复制标签失败 - 病例: {case_name}, 文件: {src_path}, 错误: {str(e)}" print(f"错误: {error_msg}") errors.append(error_msg) return False
def check_file_validity(file_path): """ 检查文件是否有效(非空且可读取) """ try: if not os.path.exists(file_path): return False, "文件不存在"
if os.path.getsize(file_path) == 0: return False, "文件为空"
# 尝试加载文件头信息 nib.load(file_path) return True, "文件有效" except Exception as e: return False, f"文件无效: {str(e)}"
training_cases = [] test_cases = [] skipped_cases = []
for i, case_name in enumerate(all_cases): case_folder = os.path.join(brats_root, case_name)
if not os.path.exists(case_folder): error_msg = f"病例文件夹不存在: {case_folder}" print(f"警告: {error_msg}") errors.append(error_msg) skipped_cases.append(case_name) continue
# 构建文件名模式 - 根据BraTS2024的命名规范 base_name = case_name # 例如: BraTS-GLI-00000-000 或 BraTS-MET-00001-000
required_files = { 't1n': f"{base_name}-t1n.nii", 't1c': f"{base_name}-t1c.nii", 't2f': f"{base_name}-t2f.nii", 't2w': f"{base_name}-t2w.nii", 'seg': f"{base_name}-seg.nii" }
# 检查文件存在性,支持.nii和.nii.gz格式 files_exist = True invalid_files = []
for key, filename in required_files.items(): file_path = os.path.join(case_folder, filename) gz_file_path = file_path + ".gz"
if os.path.exists(file_path): # 检查文件有效性 is_valid, msg = check_file_validity(file_path) if not is_valid: invalid_files.append(f"{filename}: {msg}") files_exist = False elif os.path.exists(gz_file_path): required_files[key] = filename + ".gz" # 检查文件有效性 is_valid, msg = check_file_validity(gz_file_path) if not is_valid: invalid_files.append(f"{filename}.gz: {msg}") files_exist = False else: error_msg = f"文件缺失 - 病例: {case_name}, 文件: {filename} 或 {filename}.gz" print(f"警告: {error_msg}") errors.append(error_msg) files_exist = False
if invalid_files: for invalid_file in invalid_files: error_msg = f"文件无效 - 病例: {case_name}, {invalid_file}" print(f"警告: {error_msg}") errors.append(error_msg)
if not files_exist: skipped_cases.append(case_name) continue
# 决定这个病例是用于训练还是测试(前80%用于训练) case_success = True
if i < len(all_cases) * 0.8: # 训练数据 print(f"处理训练病例: {case_name}")
# 复制图像文件 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTr_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality): case_success = False
# 复制分割标签 src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTr_folder, f"{case_name}.nii.gz") if not safe_copy_label(src_seg, dst_seg, case_name): case_success = False
if case_success: training_cases.append(case_name) print(f"训练病例 {case_name} 处理成功") else: skipped_cases.append(case_name) print(f"训练病例 {case_name} 处理失败,已跳过")
else: # 测试数据 print(f"处理测试病例: {case_name}")
# 复制图像文件 for modality, suffix in modality_mapping.items(): src_file = os.path.join(case_folder, required_files[modality]) dst_file = os.path.join(imagesTs_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality): case_success = False
# 复制测试数据的标签 src_seg = os.path.join(case_folder, required_files['seg']) dst_seg = os.path.join(labelsTs_folder, f"{case_name}.nii.gz") if not safe_copy_label(src_seg, dst_seg, case_name): case_success = False
if case_success: test_cases.append(case_name) print(f"测试病例 {case_name} 处理成功") else: skipped_cases.append(case_name) print(f"测试病例 {case_name} 处理失败,已跳过")
print(f"处理完成: {len(training_cases)} 个训练病例, {len(test_cases)} 个测试病例") print(f"跳过的病例数量: {len(skipped_cases)}")
# 写入错误日志 error_file_path = os.path.join(os.path.dirname(__file__), "error.txt") with open(error_file_path, 'w', encoding='utf-8') as f: f.write(f"BraTS2024数据转换错误报告\n") f.write(f"生成时间: {str(os.path.getctime(error_file_path)) if os.path.exists(error_file_path) else 'N/A'}\n") f.write(f"="*80 + "\n\n") f.write(f"总计处理: {len(all_cases)} 个病例\n") f.write(f"成功处理: {len(training_cases) + len(test_cases)} 个病例\n") f.write(f"跳过病例: {len(skipped_cases)} 个病例\n") f.write(f"错误数量: {len(errors)} 个错误\n\n")
if skipped_cases: f.write("跳过的病例列表:\n") for case in skipped_cases: f.write(f" - {case}\n") f.write("\n")
if errors: f.write("详细错误信息:\n") for i, error in enumerate(errors, 1): f.write(f"{i}. {error}\n") else: f.write("没有发现错误。\n")
print(f"错误日志已保存到: {error_file_path}")
# 只有成功处理的病例数量大于0时才创建dataset.json if len(training_cases) + len(test_cases) > 0: # 创建dataset.json文件 dataset_json = OrderedDict() dataset_json['name'] = "BraTS2024" dataset_json['description'] = "Brain Tumor Segmentation Challenge 2024" dataset_json['tensorImageSize'] = "4D" dataset_json['reference'] = "https://www.synapse.org/#!Synapse:syn53708249" dataset_json['licence'] = "see BraTS2024 website" dataset_json['release'] = "1.0"
# 模态信息 - BraTS2024的模态 dataset_json['modality'] = { "0": "T1n", # T1 native (非增强T1) "1": "T1c", # T1 contrast enhanced (增强T1) "2": "T2f", # T2 FLAIR "3": "T2w" # T2 weighted }
# 标签信息 - BraTS2024包含新的切除腔标签 dataset_json['labels'] = { "0": "background", "1": "NETC (Non-Enhancing Tumor Core)", # 非增强肿瘤核心 "2": "SNFH (Surrounding Non-enhancing FLAIR Hyperintensity)", # 周围非增强FLAIR高信号(水肿) "3": "ET (Enhancing Tumor)", # 增强肿瘤 "4": "RC (Resection Cavity)" # 切除腔(BraTS2024新增) }
# 训练和测试数据列表 dataset_json['numTraining'] = len(training_cases) dataset_json['numTest'] = len(test_cases)
dataset_json['training'] = [] for case in training_cases: case_dict = { "image": f"./imagesTr/{case}.nii.gz", "label": f"./labelsTr/{case}.nii.gz" } dataset_json['training'].append(case_dict)
dataset_json['test'] = [] for case in test_cases: dataset_json['test'].append(f"./imagesTs/{case}.nii.gz")
# 保存dataset.json json_file_path = os.path.join(task_folder, "dataset.json") with open(json_file_path, 'w') as f: json.dump(dataset_json, f, indent=4)
print(f"dataset.json 已保存到: {json_file_path}") print("BraTS2024数据转换完成!标签保持原始值不变:") print(" 0 - background (背景)") print(" 1 - NETC (Non-Enhancing Tumor Core) - 非增强肿瘤核心") print(" 2 - SNFH (Surrounding Non-enhancing FLAIR Hyperintensity) - 周围非增强FLAIR高信号") print(" 3 - ET (Enhancing Tumor) - 增强肿瘤") print(" 4 - RC (Resection Cavity) - 切除腔")
return task_folder else: print("警告: 没有成功处理任何病例,未生成dataset.json文件") return None
# 使用示例if __name__ == "__main__": # 设置路径 brats_root = "BraTS2024" # 您的BraTS2024数据根目录 nnunet_raw_data_base = "DATASET" # nnUNet原始数据基础目录
# 执行转换 try: result = convert_brats2024_to_nnunet(brats_root, nnunet_raw_data_base) if result: print(f"\n转换成功完成!输出目录: {result}") print("可以继续进行nnUNet的预处理和训练步骤。") print("\n注意:BraTS2024引入了新的切除腔(RC)标签(标签值4),") print("这是与之前BraTS版本的主要区别。") else: print("\n转换失败,请查看错误日志了解详细信息。") except Exception as e: print(f"程序执行失败: {str(e)}") # 即使主程序失败,也要记录错误 error_file_path = os.path.join(os.path.dirname(__file__), "error.txt") with open(error_file_path, 'w', encoding='utf-8') as f: f.write(f"程序执行失败: {str(e)}\n") print(f"错误已记录到: {error_file_path}")部分内容转载于CSDN博客:https://blog.csdn.net/chen_niansan/article/details/141527340
nnUNetv1在自己数据集上训练
https://louaq.github.io/posts/multimodal/nnunetv1在自己数据集上训练/ 最后更新于 2025-08-18,距今已过 117 天
部分内容可能已过时


