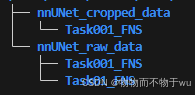
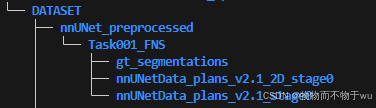
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
| import os
import shutil
import json
import nibabel as nib
import numpy as np
from collections import OrderedDict
def convert_brats2024_to_nnunet(brats_root, nnunet_raw_data_base):
"""
将BraTS2024数据集转换为nnUNet格式,保持原始标签不变
包含新的切除腔(RC)标签
Args:
brats_root: BraTS2024原始数据根目录路径
nnunet_raw_data_base: nnUNet原始数据基础目录路径
"""
errors = []
task_name = "Task001_BraTS2024"
task_folder = os.path.join(nnunet_raw_data_base, "nnUNet_raw", task_name)
imagesTr_folder = os.path.join(task_folder, "imagesTr")
imagesTs_folder = os.path.join(task_folder, "imagesTs")
labelsTr_folder = os.path.join(task_folder, "labelsTr")
labelsTs_folder = os.path.join(task_folder, "labelsTs")
for folder in [imagesTr_folder, imagesTs_folder, labelsTr_folder, labelsTs_folder]:
os.makedirs(folder, exist_ok=True)
all_cases = []
for item in os.listdir(brats_root):
case_path = os.path.join(brats_root, item)
if os.path.isdir(case_path) and item.startswith('BraTS-'):
all_cases.append(item)
all_cases.sort()
print(f"找到 {len(all_cases)} 个病例")
modality_mapping = {
't1n': '0000',
't1c': '0001',
't2f': '0002',
't2w': '0003'
}
def safe_copy_image(src_path, dst_path, case_name, modality):
"""
安全地复制图像文件,处理可能的错误
"""
try:
if not src_path.endswith('.gz'):
img = nib.load(src_path)
nib.save(img, dst_path)
else:
shutil.copy2(src_path, dst_path)
return True
except Exception as e:
error_msg = f"复制图像失败 - 病例: {case_name}, 模态: {modality}, 文件: {src_path}, 错误: {str(e)}"
print(f"错误: {error_msg}")
errors.append(error_msg)
return False
def safe_copy_label(src_path, dst_path, case_name):
"""
安全地复制标签文件,处理可能的错误
检查并记录BraTS2024的标签值分布
"""
try:
label_nii = nib.load(src_path)
label_data = label_nii.get_fdata().astype(np.uint8)
unique_labels = np.unique(label_data)
print(f"处理 {os.path.basename(src_path)},标签值: {unique_labels}")
valid_labels = {0, 1, 2, 3, 4}
unexpected_labels = set(unique_labels) - valid_labels
if unexpected_labels:
warning_msg = f"发现意外标签值 - 病例: {case_name}, 标签值: {unexpected_labels}"
print(f"警告: {warning_msg}")
errors.append(warning_msg)
label_counts = {}
for label in unique_labels:
count = np.sum(label_data == label)
label_counts[int(label)] = count
print(f" 标签分布: {label_counts}")
if not src_path.endswith('.gz'):
nib.save(label_nii, dst_path)
else:
shutil.copy2(src_path, dst_path)
return True
except Exception as e:
error_msg = f"复制标签失败 - 病例: {case_name}, 文件: {src_path}, 错误: {str(e)}"
print(f"错误: {error_msg}")
errors.append(error_msg)
return False
def check_file_validity(file_path):
"""
检查文件是否有效(非空且可读取)
"""
try:
if not os.path.exists(file_path):
return False, "文件不存在"
if os.path.getsize(file_path) == 0:
return False, "文件为空"
nib.load(file_path)
return True, "文件有效"
except Exception as e:
return False, f"文件无效: {str(e)}"
training_cases = []
test_cases = []
skipped_cases = []
for i, case_name in enumerate(all_cases):
case_folder = os.path.join(brats_root, case_name)
if not os.path.exists(case_folder):
error_msg = f"病例文件夹不存在: {case_folder}"
print(f"警告: {error_msg}")
errors.append(error_msg)
skipped_cases.append(case_name)
continue
base_name = case_name
required_files = {
't1n': f"{base_name}-t1n.nii",
't1c': f"{base_name}-t1c.nii",
't2f': f"{base_name}-t2f.nii",
't2w': f"{base_name}-t2w.nii",
'seg': f"{base_name}-seg.nii"
}
files_exist = True
invalid_files = []
for key, filename in required_files.items():
file_path = os.path.join(case_folder, filename)
gz_file_path = file_path + ".gz"
if os.path.exists(file_path):
is_valid, msg = check_file_validity(file_path)
if not is_valid:
invalid_files.append(f"{filename}: {msg}")
files_exist = False
elif os.path.exists(gz_file_path):
required_files[key] = filename + ".gz"
is_valid, msg = check_file_validity(gz_file_path)
if not is_valid:
invalid_files.append(f"{filename}.gz: {msg}")
files_exist = False
else:
error_msg = f"文件缺失 - 病例: {case_name}, 文件: {filename} 或 {filename}.gz"
print(f"警告: {error_msg}")
errors.append(error_msg)
files_exist = False
if invalid_files:
for invalid_file in invalid_files:
error_msg = f"文件无效 - 病例: {case_name}, {invalid_file}"
print(f"警告: {error_msg}")
errors.append(error_msg)
if not files_exist:
skipped_cases.append(case_name)
continue
case_success = True
if i < len(all_cases) * 0.8:
print(f"处理训练病例: {case_name}")
for modality, suffix in modality_mapping.items():
src_file = os.path.join(case_folder, required_files[modality])
dst_file = os.path.join(imagesTr_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality):
case_success = False
src_seg = os.path.join(case_folder, required_files['seg'])
dst_seg = os.path.join(labelsTr_folder, f"{case_name}.nii.gz")
if not safe_copy_label(src_seg, dst_seg, case_name):
case_success = False
if case_success:
training_cases.append(case_name)
print(f"训练病例 {case_name} 处理成功")
else:
skipped_cases.append(case_name)
print(f"训练病例 {case_name} 处理失败,已跳过")
else:
print(f"处理测试病例: {case_name}")
for modality, suffix in modality_mapping.items():
src_file = os.path.join(case_folder, required_files[modality])
dst_file = os.path.join(imagesTs_folder, f"{case_name}_{suffix}.nii.gz")
if not safe_copy_image(src_file, dst_file, case_name, modality):
case_success = False
src_seg = os.path.join(case_folder, required_files['seg'])
dst_seg = os.path.join(labelsTs_folder, f"{case_name}.nii.gz")
if not safe_copy_label(src_seg, dst_seg, case_name):
case_success = False
if case_success:
test_cases.append(case_name)
print(f"测试病例 {case_name} 处理成功")
else:
skipped_cases.append(case_name)
print(f"测试病例 {case_name} 处理失败,已跳过")
print(f"处理完成: {len(training_cases)} 个训练病例, {len(test_cases)} 个测试病例")
print(f"跳过的病例数量: {len(skipped_cases)}")
error_file_path = os.path.join(os.path.dirname(__file__), "error.txt")
with open(error_file_path, 'w', encoding='utf-8') as f:
f.write(f"BraTS2024数据转换错误报告\n")
f.write(f"生成时间: {str(os.path.getctime(error_file_path)) if os.path.exists(error_file_path) else 'N/A'}\n")
f.write(f"="*80 + "\n\n")
f.write(f"总计处理: {len(all_cases)} 个病例\n")
f.write(f"成功处理: {len(training_cases) + len(test_cases)} 个病例\n")
f.write(f"跳过病例: {len(skipped_cases)} 个病例\n")
f.write(f"错误数量: {len(errors)} 个错误\n\n")
if skipped_cases:
f.write("跳过的病例列表:\n")
for case in skipped_cases:
f.write(f" - {case}\n")
f.write("\n")
if errors:
f.write("详细错误信息:\n")
for i, error in enumerate(errors, 1):
f.write(f"{i}. {error}\n")
else:
f.write("没有发现错误。\n")
print(f"错误日志已保存到: {error_file_path}")
if len(training_cases) + len(test_cases) > 0:
dataset_json = OrderedDict()
dataset_json['name'] = "BraTS2024"
dataset_json['description'] = "Brain Tumor Segmentation Challenge 2024"
dataset_json['tensorImageSize'] = "4D"
dataset_json['reference'] = "https://www.synapse.org/#!Synapse:syn53708249"
dataset_json['licence'] = "see BraTS2024 website"
dataset_json['release'] = "1.0"
dataset_json['modality'] = {
"0": "T1n",
"1": "T1c",
"2": "T2f",
"3": "T2w"
}
dataset_json['labels'] = {
"0": "background",
"1": "NETC (Non-Enhancing Tumor Core)",
"2": "SNFH (Surrounding Non-enhancing FLAIR Hyperintensity)",
"3": "ET (Enhancing Tumor)",
"4": "RC (Resection Cavity)"
}
dataset_json['numTraining'] = len(training_cases)
dataset_json['numTest'] = len(test_cases)
dataset_json['training'] = []
for case in training_cases:
case_dict = {
"image": f"./imagesTr/{case}.nii.gz",
"label": f"./labelsTr/{case}.nii.gz"
}
dataset_json['training'].append(case_dict)
dataset_json['test'] = []
for case in test_cases:
dataset_json['test'].append(f"./imagesTs/{case}.nii.gz")
json_file_path = os.path.join(task_folder, "dataset.json")
with open(json_file_path, 'w') as f:
json.dump(dataset_json, f, indent=4)
print(f"dataset.json 已保存到: {json_file_path}")
print("BraTS2024数据转换完成!标签保持原始值不变:")
print(" 0 - background (背景)")
print(" 1 - NETC (Non-Enhancing Tumor Core) - 非增强肿瘤核心")
print(" 2 - SNFH (Surrounding Non-enhancing FLAIR Hyperintensity) - 周围非增强FLAIR高信号")
print(" 3 - ET (Enhancing Tumor) - 增强肿瘤")
print(" 4 - RC (Resection Cavity) - 切除腔")
return task_folder
else:
print("警告: 没有成功处理任何病例,未生成dataset.json文件")
return None
if __name__ == "__main__":
brats_root = "BraTS2024"
nnunet_raw_data_base = "DATASET"
try:
result = convert_brats2024_to_nnunet(brats_root, nnunet_raw_data_base)
if result:
print(f"\n转换成功完成!输出目录: {result}")
print("可以继续进行nnUNet的预处理和训练步骤。")
print("\n注意:BraTS2024引入了新的切除腔(RC)标签(标签值4),")
print("这是与之前BraTS版本的主要区别。")
else:
print("\n转换失败,请查看错误日志了解详细信息。")
except Exception as e:
print(f"程序执行失败: {str(e)}")
error_file_path = os.path.join(os.path.dirname(__file__), "error.txt")
with open(error_file_path, 'w', encoding='utf-8') as f:
f.write(f"程序执行失败: {str(e)}\n")
print(f"错误已记录到: {error_file_path}")
|